|
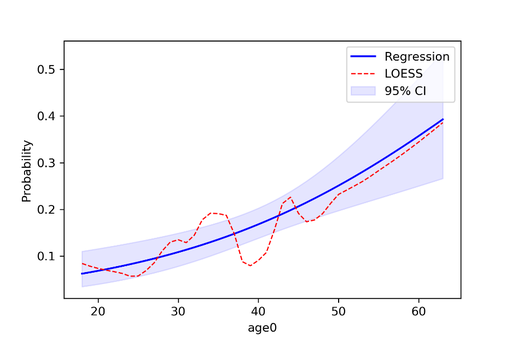
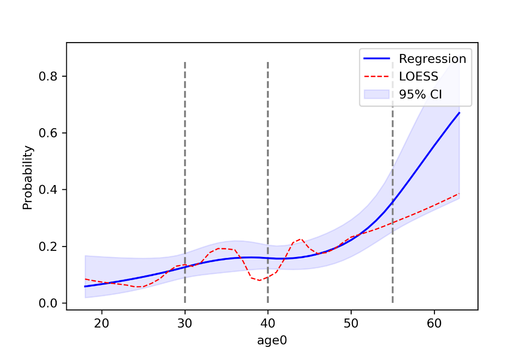
Author: Paul Zivich Python is a general computer programming language but has recently garnered popularity among data scientists with its versatility, ability to quickly process large data sets, and large library of machine learning models. I taught myself Python two years ago and while there are several Python libraries for epidemiology, I found the libraries were no longer actively maintained, did not interact with pandas (the main data management Python library), or implement causal inference methods (like inverse probability weights). To fill this gap, I created zEpid with the goal of making epidemiologic analyses in Python e-z. Functional Form AssessmentI have a few features that I especially like and will highlight them here. First is the functional form assessment. I always found coding functional form assessments to be tedious and difficult to obtain a nice-looking plot from SAS. The code I wrote creates a functional form plot and prints the model results. Below is a fully contained example import zepid as ze Which gives the following output: Warning: missing observations of model variables are dropped Generate SplinesAssessing other functional forms, creating splines, and adding points which correspond to groups of observations are also easily implementable. Since I mentioned splines, zEpid also has easy to use functionality to generate splines. The following line of code will generate a restricted quadratic spline with knots at 30, 40, and 55. Continuing with the functional form plot code from previous, we can generate another functional form plot df[['rqs0','rqs1']] = ze.spline(df,var='age0',n_knots=3,knots=[30,40,50],restricted=True) Inverse Probability WeightsLastly, zEpid has functionalities for inverse probability weights. Currently, inverse probability of treatment weights, inverse probability of censoring weights, and inverse probability of missing weights are implemented. The following block of code can be used to fit a time-fixed IPTW model. Note that we will use statsmodels to obtain the final result. Currently, zEpid only generates the weights to maintain user functionality (i.e. ability to manipulate weights for a marginal structural model). #Loading necessary packages to fit model Which gives us the following results GEE Regression Results You can visit the following website for a description on fitting a marginal structural model with an inverse probability weighted Kaplan Meier: zEpid Docs – MSM with IPW-KM For further description of the above features and others, a guide is available at http://zepid.readthedocs.io/en/latest/ Note: At the time of this blog post, we are on version 0.1.3 Download zEpidYou can download zEpid via GitHub, PyPI, or directly from the command line using pip install zepid
For an introduction to Python intended for epidemiologists, I have a guide in development at https://github.com/pzivich/Python-for-Epidemiologists Note that zEpid is distributed under the MIT license. Paul About the Author
0 Comments
Your comment will be posted after it is approved.
Leave a Reply. |
Practical solutions for conducting great epidemiology methods. Transparency in code. Attitude of constant improvement. Appreciate my stuff?
All
March 2021
|











 RSS Feed
RSS Feed