|
Author: Bailey DeBarmore Learning about a method in class, like inverse probability weighting, is different than implementing it in practice. This post will remind you why we might be interested in propensity scores to control for confounding - specifically inverse probability of treatment weights and SMR - and then show how to do so in SAS and Stata. If you have corresponding code in R that you'd like to add to this post, please contact me. A note about weighting versus multivariable regression: Effect estimate interpretations when you use weighting are marginal effect in the target population. When you adjust for covariates in a regression model, you are interpreting a conditional effect, that is, the effect of the exposure holding (conditional on) the covariates being constant. Conditional estimates are troublesome with time-varying covariates because we run into collider bias and conditioning on mediators, thus weights are preferable. In simpler situations, using weights over multivariable regression can help with convergence issues . Files to Download: .txt file with SAS and Stata code, as well as a PDF version of this post with code (perfect for students) available to download at the end of the post or at my github propensity scoresA propensity score is a predicted probability that may be used to predict exposure (or treatment) status, but can also be used for censoring or missingness. How do we use propensity scores for confounding? We can use propensity scores to generate WEIGHTS which, when applied to the final model, make the exposure independent from confounders (Figure 1B), usually by modeling the association between the exposure and confounders (instead of the main analysis where we model the outcome and exposure). Propensity scores can also control for confounding via covariate adjustment (I discourage you from this option), stratification, and matching, in addition to weighting. The 2 types of weights I'll be discussing in this post are
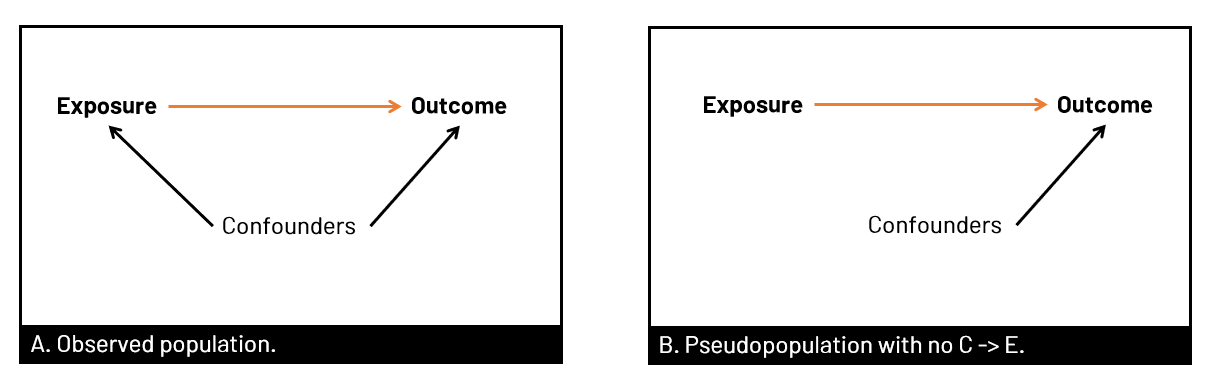 Figure 1. Panel A shows the observed population in our data set, where the relationship between exposure and outcome (orange) is confounded by well, confounders. In B, we have removed the arrow from confounders to exposure. We can remove the arrow in several ways, including using propensity scores (of various types) to create a pseudopopulation where the exposure and confounder are no longer associated. 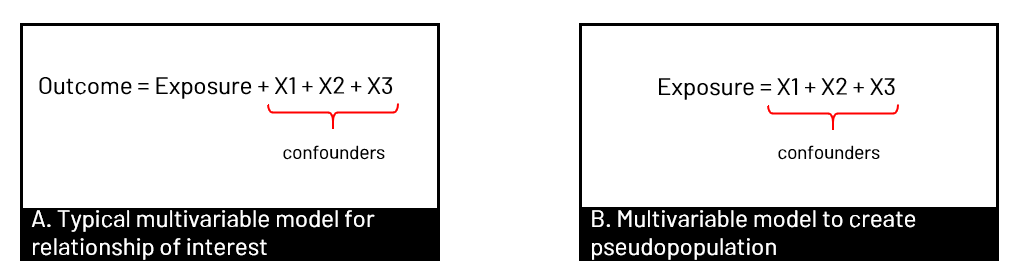 Figure 2. Panel A shows the usual multivariable model we run in our analyses - to estimate the association of the exposure with the outcome, controlled for confounders. When we want to use propensity scores, we first create the weights that we will later use in our final model by modeling the association of the confounders with the exposure - so we can remove that arrow. SMRWhen you use SMR weights, you're estimating the average treatment effect in the treated (ATT). In other words, you estimate the effect had the exposed group been exposed, versus the exposed group been unexposed. The pseudopopulation that you create has a covariate (confounder) distribution equal to that observed in the exposed group. You can also generate your SMR weights where the unexposed group is the target of interest, and model the effect had the unexposed group been unexposed versus the unexposed group been exposed. 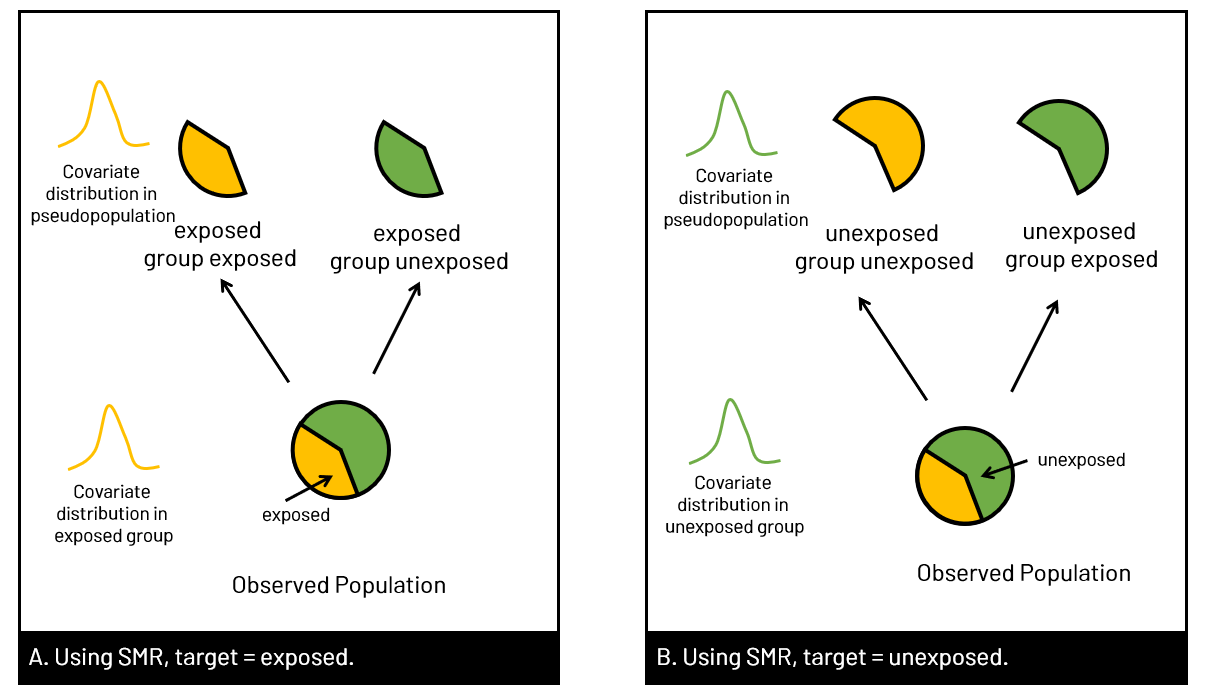 Figure 3. In Panel A, the target group is the exposed group, so we use SMR to model had the exposed group been exposed versus unexposed, with the covariate distribution of the exposed group (yellow). In Panel B, we use the covariate distribution of the unexposed group (green), and model had unexposed group been unexposed vs exposed. 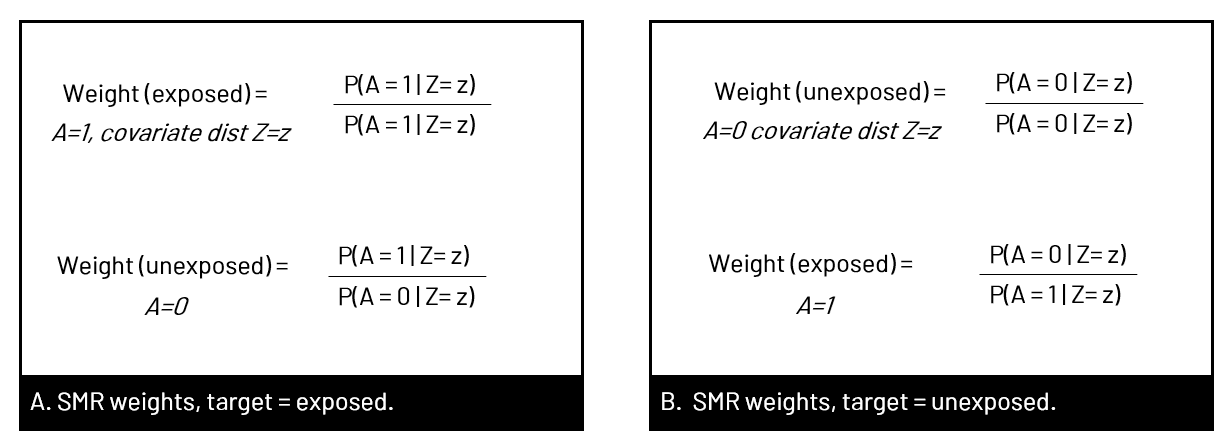 Figure 4. Equations to calculate the SMR weights for given target population corresponding to Figure 3 examples. A indicates exposure status (=1 for exposed, =0 for unexposed). The numerator is the probabilitiy of exposure given the covariate distribution of the target group, and the denominator is the probability of being assigned to what was observed. You can see that in panel B, it switches when the target group is unexposed. coding SMR in SASTo use SMR, you'll be generating the probabilities for the numerator and denominator, and then creating the weight (numerator/denominator) and applying it in the appropriate statement.
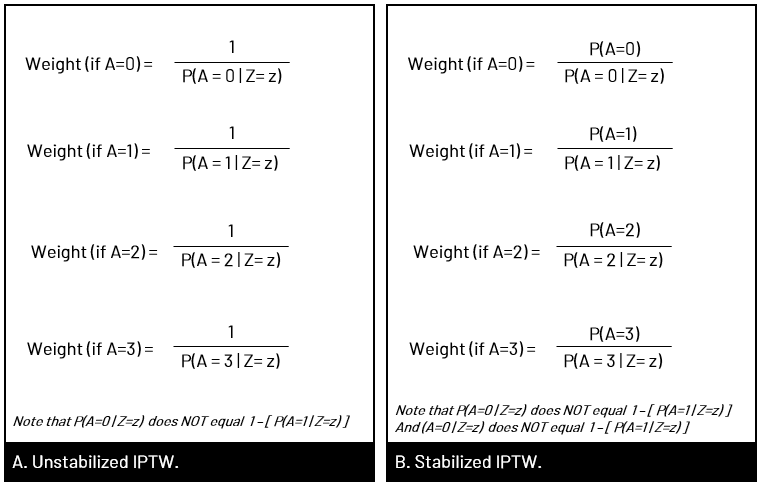
What is &let? So that you can easily adapt my code, I use &let statements at the beginning of my code blocks. After the equals sign, you would replace <data> with your dataset name, <exposure> with your exposure variable, and <outcome> with your outcome variable. The code as written will then run with those chosen variables. Note that you do need to replace <covariates> in the model statement with your confounders. If you don't want to use &let statements, simply go through the code and anywhere you see &<text>, replace both the & and the text with your regular code. coding SMR in StataThe approach to calculating SMR is different in Stata. You'll be using the built in teffects command (reference manual [TE] here) and using options to specify SMR versus IPW. Since using SMR gives us the average treatment effect in the treated, we'll use the option atet (average treatment effect on the treated) instead of ate (average treatment effect) which we'll use for IPTW. ****************************************** inverse probability of treatment weights (IPTW)In contrast to SMR weights, when you use IPTW weights you are estimating the average treatment effect (ATE), that is the treatment effect in a population with covariate distribution equal to the entire observed study population, not just the exposed or unexposed. Thus, you're modeling the complete counterfactual. In other words, you're estimating the effect had the entire population been exposed versus the entire population been unexposed. You can use unstabilized or stabilized IPTW in your final model. Choosing one over the other doesn't change your ultimate interpretation but affects the width of your confidence interval (wider when you use unstabilized). 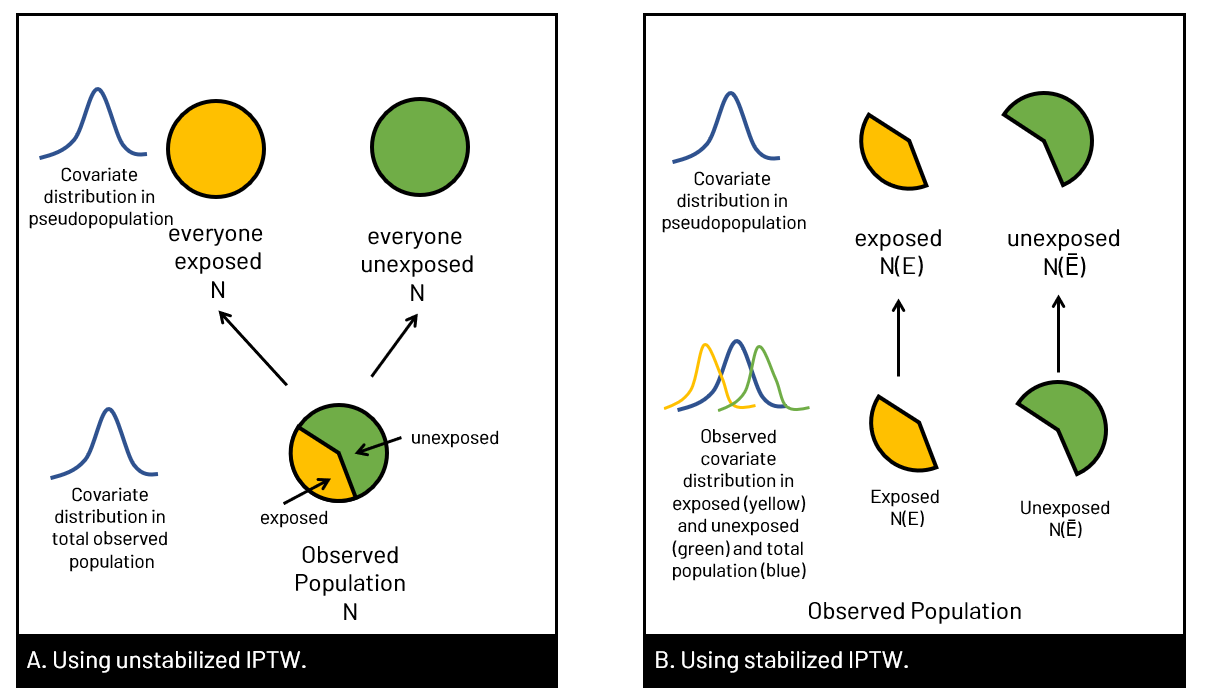 Figure 5. Panel A shows what happens in the pseudopopulation when we use unstabilized weights. We duplicate our N, creating 2 new populations with the covariate distribution of the entire observed population, but had everyone been exposed versus everyone unexposed. In B we apply the covariate distribution within strata of exposure, maintaining the same N. (E with a bar (macron) indicates unexposed).  Figure 6. In Panel A we have the equation for unstabilized IPTW for exposed and unexposed, and in Panel B, the stabilized IPTW for exposed and unexposed. You'll see additional notes that show you that you can also calculate the weight components for unexposed by subtracting the exposed probabilities from 1. You'll see this used in our code below.
You can see in Figure 5 versus Figure 3 that we are using different covariate distributions. With SMR where we match the covariate distribution of the exposed or unexposed group (whichever is target). This is why IPTW generates an average treatment effect while SMR generates an average treatment effect in the treated. coding IPTW in SAS****************************************** coding IPTW in StataThe approach to calculating IPTW is different in Stata than SAS. You'll be using the built in teffects command (reference manual [TE] here) and using options to specify IPTW. Since using IPTW results in the average treatment effect overall, we'll use option ate (average treatment effect) instead of option atet (average treatment effect on the treated) that we use for SMR. ****************************************** UPDATE: Feb 22 2021 If your treatment (exposure) is categorical you'll need to change things up a bit. I'm writing this blurb in response to a comment below, and in response to this topic coming up several times in just the past week! In your software of choice, you need to calculate the probability of being treated given your covariates, save those probabilities, and then assign the probability given each observations observed status. In SAS, you can use code like the sample below (thanks, Paul!) which uses a generalized logit model for a treatment variable <treatment> with multiple categories. The output data set will have probabilities for every value of treatment. For example, a person with treatment=1 will have values for treatment=0,1,2,3 so you need to do some more coding to tell the computer you only want the predicted value for treatment if treatment=treatment. PROC LOGISTIC DATA=...; In Stata, you can run mlogit to estimate the probability of <treatment> adjusted for variables <vars> and then use the postestimation commands, predict, to create a denominator variable for each value of treatment (0, 1, 2, 3). You use the option "outcome" to tell Stata this. mlogit treatment vars Then create your weight variable, iptw below. gen iptw=. If you want to create stabilized weights, you can run a tab to get the proportion in each category, and then calculate your weights. For example, let's say my groups are distributed as P(A=0)=0.6, P(A=1)=0.14, P(A=2)=0.2, and P(A=3)=0.06, I would make a stabilized iptw variable, siptw, below. gen siptw=. Download resources here, or get the most up-to-date versions on my GitHub
Suggested citationDeBarmore BM. “Coding IPW and SMR in SAS and Stata”. 2019. Updated 2021. Retrieved from http://www.baileydebarmore.com/epicode/calculating-ipw-and-smr-in-sas About the author
15 Comments
Ken
2/5/2021 01:34:43 am
Hi Bailey. Really liked your post! I have a quick question: For IPTW, I wonder if the SAS codes can also be accommodated for an exposure with more than two groups. Say if x (exposure) can be 1, 2, or 3, then should we specify the stabilizing weight to be sw=(1-n)/(1-d) for x=1 (the referent group) and sw = n/d for x=2 and x=3 (the rest two groups)?
Reply
Bailey
2/5/2021 10:28:19 am
Hi Ken,
Reply
Ken
2/5/2021 04:05:59 pm
Thanks Bailey and Paul for helping me on this! I found a SAS document that seems to suggest a similar solution to what you described: https://support.sas.com/resources/papers/proceedings/proceedings/forum2007/184-2007.pdf
Reply
Ann
2/26/2021 12:55:54 am
Hi Bailey,
Reply
Bailey
2/26/2021 09:27:58 am
Great question, Ann! I've added some code to the Stata portion that specifies how to do this. Hope that helps!
Reply
Ann
2/26/2021 11:01:22 am
Thank you I just saw that! I did have a follow-up question, when you mention stabilizing the iptw, is it the same as normalizing the iptw? Performed as follows:
Bailey
2/26/2021 11:34:39 am
Hi Ann,
Reply
Diego
4/9/2021 07:00:37 pm
Thanks for the information. It's really useful.I was wondering if the process is the same for IPW for attrition?
Reply
Bailey
5/24/2022 10:19:01 am
Hi Diego, thanks for the comment. The process would be the same or similar for IPW of attrition, you would replace your predictors of treatment with predictors of attrition. - Bailey
Reply
Joseph
4/28/2022 12:19:27 pm
Baily:
Reply
Bailey
5/24/2022 10:18:25 am
Hi Joseph, thanks for the comment (and the heads up about the typo - fixed now).
Reply
Matthew
8/26/2022 12:26:43 pm
Hi Bailey, this is an amazing post about IPTW and just what I have been looking for.
Reply
Bailey
8/30/2022 10:47:27 am
You may find the command "tebalance" helpful. This article discusses it (scroll down until you see the balance plots).
Reply
Selina
2/6/2023 05:37:32 am
Hi Bailey,
Reply
Val
9/14/2023 04:36:08 pm
I have data where the unexposed group is much larger than the exposed group, so the SMR weights are very small for the unexposed group. Is it valid to normalize the SMR weights for the unexposed group to have a mean of 1 (i.e. divide the SMR weights by the mean for the group). That makes the sum of all the weights similar to the total study population, but does it still mean that I am estimating ATT?
Reply
Your comment will be posted after it is approved.
Leave a Reply. |
Practical solutions for conducting great epidemiology methods. Transparency in code. Attitude of constant improvement. Appreciate my stuff?
All
March 2021
|
||||||||||||||||||||||










 RSS Feed
RSS Feed